In-depth analysis of complex genetic variants
S E Q U E G E N I C S® is providing researchers and clinicians with comprehensive solutions and personalized support. We empower them to attain a unique in-depth comprehension of the genetic makeup underlying diseases, a feat made possible only through the utilization of the most sophisticated DNA sequencing technologies.
In addition to error correction, genome mapping and variant calling, our processes implement both simple and complex variant interpretation algorithms to determine their potential deleterious effect.
We integrate these pipelines into a cloud platform that facilitates the navigation of complex data sets in a highly efficient manner.
We delve deeper than ever before into the genetic composition of patients to find answers to previously unsolvable questions.
With long-read sequencing, you will be able to analyze more complex genomic structures than short-read sequencing allows.
We build unique, propietary pipelines on top of the two major long-read technologies
At Sequegenics® we not only provide analysis services but also train clinicians and researchers in the unique in-depth understanding of genetics made possible by the most sophisticated sequencing technologies.
Achieve more effective results in less time and at a lower cost
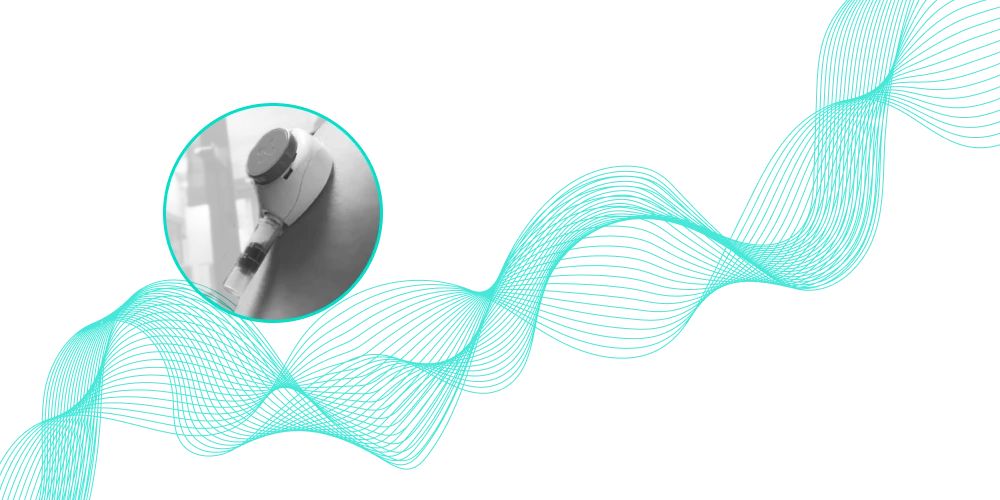
Obtain a sequenced long-read genome for less than 500 µL of blood collected at home.
Other providers | Sequegenics® | |
|---|---|---|
High-molecular weight gDNA extraction included | ||
Average Read Length | ⁓ 15kb | ⁓ 25kb |
Sequencing Coverage | ~30x | ~50x |
Turnaround Time | > 2 months | < 3 weeks |
Variant interpretation included |

Write to us at contact@sequegenics.com and someone from our team will contact you.